do i need to autoclave dialysis buffer|cellulose dialysis : private label Change the dialysis buffer and dialyze for another 1 to 2 h. Change the dialysis buffer and dialyze overnight at 4°C. Note: For best results, use a volume of dialysis buffer (dialysate) that is at least 200-fold greater than the sample volume. To conserve dialysate with large-volume devices, such as the Slide-A-Lyzer dialysis flasks, a 5-fold . $545.00
{plog:ftitle_list}
Sterilization is a critical aspect of veterinary care, safeguarding animals from .
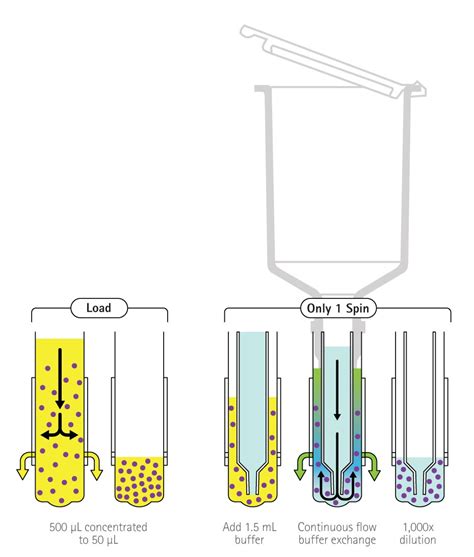
Dialysis is a separation technique that facilitates the removal of small, unwanted compounds from macromolecules in solution by selective and passive diffusion through a semi-permeable membrane. A sample and a buffer solution (called the dialysate, usually 200 to 500 times the .Thermo Scientific dialysis devices are available in a variety of dialysis options for the rapid and trouble-free dialysis of sample volumes ranging from 10 μL to 250 mL. Unlike traditional dialy.Dialysis is a classic separation technique that facilitates the removal of small, unwanted compounds from macromolecules in solution by selective diffusion through a semi-permeable .
Change the dialysis buffer and dialyze for another 1 to 2 h. Change the dialysis buffer and dialyze overnight at 4°C. Note: For best results, use a volume of dialysis buffer (dialysate) that is at least 200-fold greater than the sample volume. To conserve dialysate with large-volume devices, such as the Slide-A-Lyzer dialysis flasks, a 5-fold . You can do everything without filter sterilizing but you would also need to bake/ autoclave anything that comes in contact with the water when you make the buffer, or you can buy pre-sterile tubes .What dialysis buffer should I use for the protein subsequently to be used for ki - What decides the composition of the dialysis buffer? (reply: 1) . which buffer need to autoclave, which can not - (reply: 1) preparation of solutions that need pH adjustment - (reply: 3)
My His-tagged purified protein contains 10mM trisCl, 300mM NaCl and 200mM imidazole at which the protein was eluted at pH 8.0. What should be ideal dialysis buffer composition to remove imidazole?Why do I have to add beta-mercaptoethanol (beta-ME) to lysis Buffer RLT of the RNeasy Kits? When working with RNA, care must be taken to avoid degradation by RNases, which are extremely stable and active. Intracellular RNases are released during the lysis step of the RNA isolation procedure and must be rapidly and thoroughly inactivated to .
the rails inside the autoclave to unload the shelf (Fig #7). NEVER!! load the autoclave without using the cart to support the shelf. Be very careful to not touch anything without gloves on, since all surfaces are HOT!! (3) Pancoe Autoclave Room (4) Pancoe autoclave (5) Program Screen (6) Jacket PressureMy general rule is that buffer volume has to be 10X-20X the sample. Dialysis duration (O/N rather than few hours), temperature (I assume yuo operate at about 4°C) and buffer changes affect the .I am using CTAB method (Naser M. Salameh et al., 2014) for fungal DNA isolation and purification. During reagents preparation I am not sure weather I should autoclave 2X CTAB and Wash buffer.
protein dialysis buffer settings
autoclave separately. for sugar solution 10 minutes, psi 10, and temperature 121 and for medium 15 minutes 15psi ad 121 temperature. after the autoclaving mix both solution and pour into Petri plate.ChIP Dialysis buffer -Rabbit 1000 ml 50 mM Tris-Cl pH 8.0 1 M 50 ml 0.2% Sarkosyl 20% 10 ml 2 mM EDTA 0.5 M 4 ml ddH2O 30.2g PIPES, 17 926 ml ChIP Dialysis buffer -Mouse 1000 ml 50 mM Tris-Cl pH 8.0 1 M 50 ml 2 mM EDTA 0.5 M 4 ml ddH2O 946 ml ChIP Wash buffer-Rabbit 1000 ml 100 mM Tris, pH 9.0 1 M 100 mlAllow the solution to cool to room temperature before making final adjustments to the pH. Adjust the volume of the solution to 1 L with H 2 O. Dispense into aliquots and sterilize by autoclaving.. If the 1 M solution has a yellow color, discard it and obtain Tris of better quality.Hello Linda, I also recommend use a commercial source of stock buffers, like 5M NaCl, NP40, and nuclease free water also to prepare buffer for RNA-IP purpose. Autoclave somehow may not inactive .
Glycerol can perfectly withstand autoclaving. However, for practical purposes, I prefer to autoclave a 60 % (v/v) glycerol solution, prepared by weighing glycerol to avoid problems due to glycerol .Adjust the final volume to 1000 ml (1Lt) and autoclave the solution. The solution is ready to use. All the best. ;) . Yes, you can put it to autoclave or filter it if you need sterile buffer.
Buffer Calculations: Formula and Equations. Molar solution equation: desired molarity × formula weight × solution final volume (L) = grams needed Percentage by weight (w/v): (% buffer desired / 100) × final buffer volume (mL) = g of starting material needed Henderson-Hasselbach equation: pH = pKa + log [A-]/[HA] The Henderson-Hasselbalch equation enables determination of a .
Dialysis of 100 µL of small compounds, 500–1,500 daltons, against a saline solution will be ~50% complete in 2–4 hours or 99%+ complete after overnight when dialyzing against ~1 liter of buffer. Dialysis will proceed faster with more frequent buffer changes.Dialysis of 100 µL of small compounds, 500–1,500 daltons, against a saline solution will be ~50% complete in 2–4 hours or 99%+ complete after overnight when dialyzing against ~1 liter of buffer. Dialysis will proceed faster with more frequent buffer changes. To summarize, each purification strategy has its buffer recommendations. First, you need to choose the purification strategy and after that you can select the most adapted buffer. 3# Selection of buffer Components. Each buffer has a range of pH in which its buffered power is efficient. To be sure that you have chosen the right buffer for a .
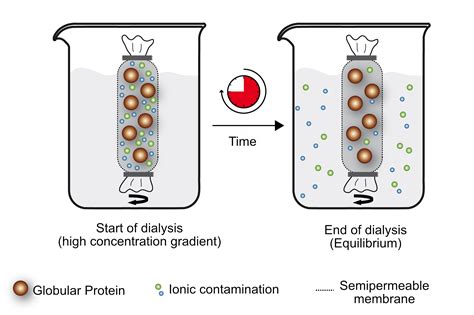
A dialysis membrane is a semi-permeable film (usually a sheet of regenerated cellulose) containing various sized pores. Molecules larger than the pores cannot pass through the membrane but small molecules can do so freely. In this manner, dialysis may be used to perform purification or buffer exchange for samples containing macromolecules.Dialysis of 100 µL of small compounds, 500–1,500 daltons, against a saline solution will be ~50% complete in 2–4 hours or 99%+ complete after overnight when dialyzing against ~1 liter of buffer. Dialysis will proceed faster with more frequent buffer changes.
TE buffer: 10 mM Tris-Cl (pH, usually 7.6 or 8.0) 1 mM EDTA (pH 8.0) Use concentrated stock solutions to prepare. If sterile water and sterile stocks are used, there is no need to autoclave. Otherwise, sterilize solutions by autoclaving for 20 minutes. Store the buffer at room temperature.The dissolved mucin was dialyzed overnight against three changes of 5 L phosphate buffer using dialysis membrane with a 2300 molecular weight cut off (Spectra/Por 7 Dialysis Membrane; Spectrapor).
protein dialysis buffer
Whatever you do or don't do, just keep a negative control or non-template control in your experiments. That will directly solve your query as when and what to autoclave. No, you don't need to autoclave. But be aware that some protein quatification methods find very hard to measure protein concentrations in such a high urea concentrations.I had a coulple of troubles using 6M urea with 2 M thiourea for extraction buffer..my BCA anaylis went crazy...all I could measure was urea..Dialysis of 100 µL of small compounds, 500–1,500 daltons, against a saline solution will be ~50% complete in 2–4 hours or 99%+ complete after overnight when dialyzing against ~1 liter of buffer. Dialysis will proceed faster with more frequent buffer changes.Dialysis of 100 µL of small compounds, 500–1,500 daltons, against a saline solution will be ~50% complete in 2–4 hours or 99%+ complete after overnight when dialyzing against ~1 liter of buffer. Dialysis will proceed faster with more frequent buffer changes.
Dialysis of 100 µL of small compounds, 500–1,500 daltons, against a saline solution will be ~50% complete in 2–4 hours or 99%+ complete after overnight when dialyzing against ~1 liter of buffer. Dialysis will proceed faster with more frequent buffer changes.
is the real mcat harder than the practice tests
is the real mcat harder than the practice tests reddit
Autoclavable materials come out completely sterilized free of infectious bacteria and can either be thrown away like regular municipal waste or reused for the treatment of patients.
do i need to autoclave dialysis buffer|cellulose dialysis